Today is the start of regular release of Wash U Epigenome Browser source code. The first version is available here: http://epigenomegateway.wustl.edu/source/ This link is also accessible from browser home page.
Inside this directory there's currently one compressed file marked as "v1". Download the file and get all the working code of the Browser. As stated on the browser web site, the code is free for non-commercial use.
The contents in the downloaded file are a freeze of my working directory. Most of the files are text files. Only exception is a few key documents with suffix ".dia". The software Dia is required to open them.
About the version number: an integer starting from 1 as of this first release, to be incremented by 1 at each new release. This is my personal tribute to the prestigious UCSC Kent Source Tree, the fundamental basis of my work, which is now at v265 over its bi-weekly release cycle. Currently we're yet to move to GitHub because we don't have any plan to branch out from existing code.
Each release will be accompanied by a blog post to explain bug fixes/updates/new features.
===
UPDATES AND FIXES
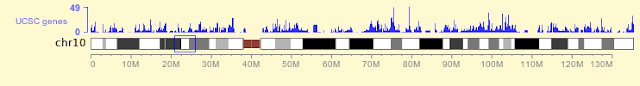
Gene tracks has been updated and an error with gene information table in database is corrected (the chromosome name field was defined as char(10) so chromosome names longer than 10 characters was truncated and led to errors).
The centralized gene symbol and description table is discarded, and now each gene type will have its own symbol-description table.
Transposable element tracks were added for fruit fly and Arabidopsis.
[ Human ]
the UCSC Gene track, RefSeq gene track, Ensembl gene track, GENCODE gene track source data were retrieved from UCSC Genome Browser ftp site on April 26th, 2012.
[ Mouse ]
the UCSC Gene track, RefSeq gene track, Ensembl gene track data were retrieved from UCSC Genome Browser ftp site on April 26th
[ Zebrafish ]
the RefSeq and Ensembl gene tracks source data were retrieved from UCSC Genome Browser ftp site on April 26th
[ Fruitfly ]
the RefSeq, Ensembl, and FlyBase gene tracks and RepeatMasker source data were retrieved from UCSC Genome Browser ftp site on April 26th
[ Arabidopsis ]
the TAIR gene track data and transposon information was retrieved from TAIR ftp site on April 27th
Inside this directory there's currently one compressed file marked as "v1". Download the file and get all the working code of the Browser. As stated on the browser web site, the code is free for non-commercial use.
The contents in the downloaded file are a freeze of my working directory. Most of the files are text files. Only exception is a few key documents with suffix ".dia". The software Dia is required to open them.
About the version number: an integer starting from 1 as of this first release, to be incremented by 1 at each new release. This is my personal tribute to the prestigious UCSC Kent Source Tree, the fundamental basis of my work, which is now at v265 over its bi-weekly release cycle. Currently we're yet to move to GitHub because we don't have any plan to branch out from existing code.
Each release will be accompanied by a blog post to explain bug fixes/updates/new features.
===
UPDATES AND FIXES
Gene tracks has been updated and an error with gene information table in database is corrected (the chromosome name field was defined as char(10) so chromosome names longer than 10 characters was truncated and led to errors).
The centralized gene symbol and description table is discarded, and now each gene type will have its own symbol-description table.
Transposable element tracks were added for fruit fly and Arabidopsis.
[ Human ]
the UCSC Gene track, RefSeq gene track, Ensembl gene track, GENCODE gene track source data were retrieved from UCSC Genome Browser ftp site on April 26th, 2012.
[ Mouse ]
the UCSC Gene track, RefSeq gene track, Ensembl gene track data were retrieved from UCSC Genome Browser ftp site on April 26th
[ Zebrafish ]
the RefSeq and Ensembl gene tracks source data were retrieved from UCSC Genome Browser ftp site on April 26th
[ Fruitfly ]
the RefSeq, Ensembl, and FlyBase gene tracks and RepeatMasker source data were retrieved from UCSC Genome Browser ftp site on April 26th
[ Arabidopsis ]
the TAIR gene track data and transposon information was retrieved from TAIR ftp site on April 27th