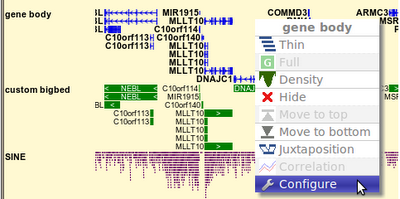
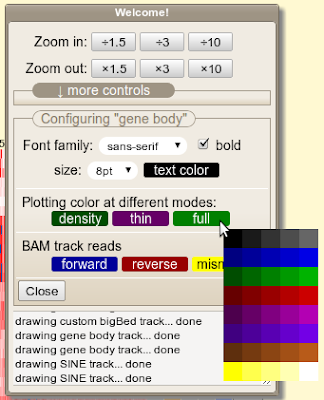
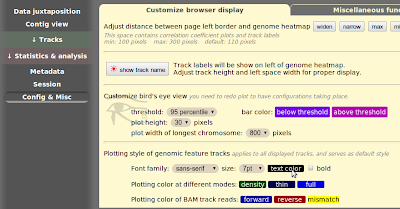
One shortcoming of "Gene set view" function is that you can't view too many genes at one time, say, >200 genes. In order to overcome this, the "bigBed file generator" is written, which allows you to submit arbitrary number of genes/coordinates and make a bigBed file which gives you wide range of browsing options once it's up as a custom track.
The bigBed file generator function interface can be found at "Miscellaneous functions" tab:
Enter a list of gene names or coordinates into the text area and you are good to go! Of course you can change your idea and select different part of genes to be used by clicking "Change" button:
The "gene part selector" panel will appears:

Click button to generate bigBed file, file URL will be given:
Finally, you can click button under the file URL and put it up as custom bigBed track instantly:
Have fun browsing!